Wet lab
Central national infrastructure for high-density data
1. Laboratories
Central Next Generation Sequencing laboratory, location Maribor
The laboratory offers ELIXIR-SI owned state of the art NGS and other high-throughput laboratory equipment, tools and services.
Laboratory equipment manager: research assistant Jan Gojznikar. Contact: elixir.mb@mf.uni-lj.si
Laboratory for single cell analysis, location Ljubljana
The laboratory offers ELIXIR-SI owned state of the art equipment, tools and services for single cell analysis.
Coordinator of the use of research infrastructure in both laboratories: research associate dr. Polonca Ferk. Contact: elixir@mf.uni-lj.si
Other laboratories
2. Research equipment
ELIXIR-SI RI-SI-2 project Research equipment |  |
Central Next Generation Sequencing laboratory, location Maribor

The nCounter® SPRINT Profiler (manufactured by NanoString Technologies, Inc.) allows simultaneous quantitative analysis of up to 800 nucleic acid sequences (DNA and RNA) or proteins in a sample (or simultaneous quantitative analysis of up to 200 nucleic acid sequences in 4 samples). Fluorescently labeled probes bind to specific nucleic acid sequences in the sample, while unbound probes are removed during sample preparation.
Using a microscope and computer image analysis, the system determines the type and number of bound probes and thus enables the quantitative analysis of nucleic acids and proteins. The advantage of the system is the quick and easy preparation of the sample for analysis, while no amplification, transcription into cDNA and library preparation is needed. It also allows analysis of complex samples such as FFPE (Formalin-Fixed Paraffin-Embedded) samples, tissue samples, lysates and fluids.
Location: ELIXIR-SILVR (KRVS) unit.
 The SeqStudio Genetic Analyzer is the fourth generation of 4 capillary, electrophoresis (CE) so-called Sanger sequencers. SeqStudio is an automated capillary electrophoresis system for nucleic acid analysis. It automatically performs sampling, polymer exchange, DNA separation and simultaneous detection of fluorescence in 4 capillaries. The system supports both sequential and fragment analysis on a plate (96-well) or in tubes (8-set) at the same time. Additionally, it allows the addition of new samples during the analysis. The work analysis program can be prepared on a computer, on the device itself (large touch screen), downloaded via WI-FI, LAN communication.
The SeqStudio Genetic Analyzer is the fourth generation of 4 capillary, electrophoresis (CE) so-called Sanger sequencers. SeqStudio is an automated capillary electrophoresis system for nucleic acid analysis. It automatically performs sampling, polymer exchange, DNA separation and simultaneous detection of fluorescence in 4 capillaries. The system supports both sequential and fragment analysis on a plate (96-well) or in tubes (8-set) at the same time. Additionally, it allows the addition of new samples during the analysis. The work analysis program can be prepared on a computer, on the device itself (large touch screen), downloaded via WI-FI, LAN communication.
SeqStudio supports the same reagents as other Applied Biosystems multi-capillary sequencers (family 3500 and 3730, old systems 310 and 3130). Chemistry of BigDye ™ Terminator 1.1 Kit and BigDye ™ Terminator 3.1 Kit terminator are used for sequence analysis. Several dye chemistry is supported to perform fragment analysis, up to and including the last six dye chemistry.
The main applications are: plasmid sequencing, analysis in oncology (mutation confirmation-orthogonal technology), species identification, CRISPR-Cas9 analysis – genome editing (diagnostics, pharmacy), forensic analysis, SNP analysis (SNAP shot genotyping), multiplex ligation ( MLPA; CNV) and other analyzes in pharmacy, diagnostics, environmental protection, microbiology.
In addition to classical sequential and fragment analyzes, the system enables various analyzes useful in diagnostics from HLA, CF, HIV, analysis of bacterial and fungal communities, comparative sequencing, validation of certain regions.
Location: ELIXIR-SILVR (KRVS) unit.
Equipment reservation
In case the desired date is not available, please write to us at info@elixir-slovenia.org

The upright confocal laser scanning microscope is equipped with a motorized microscope table which enables scanning along the x and y axes and focusing along the z axis.
It contains 10x, 40x and 63x objectives, 448 nm, 488 nm, 552 nm and 638 nm wavelength lasers and two scanning detectors, one of which is a super sensitive GaAsP detector. To ensure optimal conditions for multicolor imaging, the detectors provide a freely adjustable wavelength detection bandwidth.
The software allows time-lapse imaging in xyz coordinates, uncoupling the spectra of individual fluorescent dyes and motorized scanning of multiple regions of interest. The main advantage of the microscope is the large working distance (25 cm) between the motorized microscope table and the target focal plane, which allows in-vivo observation of large samples, such as plants.
Location: ELIXIR-SILVR (KRVS) unit.
The RIEGL miniVUX-2UAV is an extremely light laser scanner designed specifically for integration with drones. The sensor enables a pulse repetition rate of 100 kHz to 200 kHz, ie with 200,000 measurements per second, with a 360-degree field of view. This creates a dense dot pattern on the ground and thus allows good resolution capture of small objects. Spatially accurate data is provided by three GPS / GNSS sensors in the Trimble APX-15 UAV (V2) inertial system. The latter allows direct georeferencing with data collected by sensors mounted on the drone. The DJI Matrix 600 Pro is a six-rotor drone and is equipped with hardware and software support for flying missions. Autopilot A3 Pro uses diagnostic algorithms to compare data from all three GNSS units and, together with dedicated vibration damping, ensures a stable flight.
Typical applications: Agriculture and forestry (e.g. plant canopy architecture and phenotyping), mapping of glaciers into snowfields, archaeology and cultural heritage documentation, construction site monitoring, landslide monitoring.
 The NextSeq2000 system enables innovative and highly efficient biological systems research and works on various next-generation sequencing (NGS) applications. Applications include small whole-genome (≤5 Mb; bacteria, viruses, and other microbes), exomes, amplicons, target enrichment, total or targeted RNA sequencing, Chip- and ATAC-Seq, methylation, ctDNA, mRNA, and single-cell RNA sequencing or targeted gene panels sequencing.
The NextSeq2000 system enables innovative and highly efficient biological systems research and works on various next-generation sequencing (NGS) applications. Applications include small whole-genome (≤5 Mb; bacteria, viruses, and other microbes), exomes, amplicons, target enrichment, total or targeted RNA sequencing, Chip- and ATAC-Seq, methylation, ctDNA, mRNA, and single-cell RNA sequencing or targeted gene panels sequencing.
The system is based on the patterned flow cell technology, allowing significantly increased data processing capacity compared to other NGS systems. At the same time, faster sequencing with the same accuracy is provided by two-channel synthesis sequencing (SBS). The system takes advantage of an integrated cartridge that includes reagents, fluidics, and the waste holder, simplified library loading and flow cell insertion as well as simplified instrument use. The NextSeq2000 generates enormous amounts of DNA data; ultra high-density flow cells enable tunable outputs from 60 to 360 Gb. Fast and accurate analysis performed by DRAGEN Bio-IT platform is suitable for novice and expert NGS users alike.
Location: ELIXIR-SILVR (KRVS).
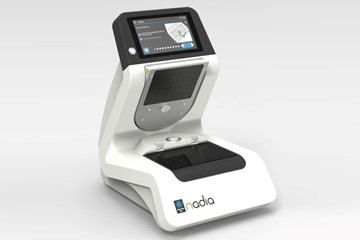 The Nadia instrument (Dolomite Bio) is an automated microfluidic system for single-cell research that encapsulates up to 8 samples, in parallel, in under 20 minutes. During the run, single cells are encapsulated in microfluidic droplets with mRNA capture beads. Over 50,000 single cells can be captured per cartridge in a run. Nadia Instrument contains three independent ultra-smooth pressure pumps, system for sample chilling and system for gentle cell agitation prior encapsulation. System can be used to encapsulate single cells or nuclei from eukaryotic sample types, such as tissues, blood, biopsies, tumours, yeasts or even plant protoplasts.
The Nadia instrument (Dolomite Bio) is an automated microfluidic system for single-cell research that encapsulates up to 8 samples, in parallel, in under 20 minutes. During the run, single cells are encapsulated in microfluidic droplets with mRNA capture beads. Over 50,000 single cells can be captured per cartridge in a run. Nadia Instrument contains three independent ultra-smooth pressure pumps, system for sample chilling and system for gentle cell agitation prior encapsulation. System can be used to encapsulate single cells or nuclei from eukaryotic sample types, such as tissues, blood, biopsies, tumours, yeasts or even plant protoplasts.
Location: unit ELIXIR-SILVR (KRVS).
 The MARVIN ProLine seed analyser (MARViTECH GmbH) is a measuring system for daily laboratory use of samples of different kind of seeds (cereals, vegetables, flowers, oil-seeds etc.) of grain sizes from 0,8mm.
The MARVIN ProLine seed analyser (MARViTECH GmbH) is a measuring system for daily laboratory use of samples of different kind of seeds (cereals, vegetables, flowers, oil-seeds etc.) of grain sizes from 0,8mm.
It enables:
– measuring of total number of the sample, thousand grain weight (TGW), grain dimension (length, width, area), determination of dark or bright seeds (for flat seeds);
– determination of size fractions (length, width, area), hectolitre weight (approximately), single seed data.
The measuring time is very fast (< 3 sec), counting accuracy is at least 99%, a large seed tray enables measurements of a high number of seeds. Preconfigured connecting and controlling of electronic scales, printer and barcode-reader is possible. All measuring data can be exported in *.csv, *.txt or *.xls format or to database.
 The SimpliAmp™ Thermal Cycler (Thermo Fisher Scientific, Inc.) is a small and accurate thermal cycler, specifically designed for the amplification of nucleic acids using the polymerase chain reaction (PCR) process. The user interface includes a touchscreen with a graphical display that shows the time, status, and temperature for each step in a PCR run. A touchscreen keypad allows you to enter information into fields on the display screen. Equipment has a simple lid design, which is easy to use and designed to help protect your samples. The equipment is compatible with the Applied Biosystems™ Thermal Cycler Fleet Control Software. Key features and benefits of the thermal cycler are optimizing your PCR with the help of the Veriflex™ blocks (which provide 3 independent temperature zones, for precise temperature control in each step of PCR process), remote access to the system through a mobile application PCR Essentials, transfering data through USB port and simulation modes library, which allows us to run an experiment using the same ramp rates.
The SimpliAmp™ Thermal Cycler (Thermo Fisher Scientific, Inc.) is a small and accurate thermal cycler, specifically designed for the amplification of nucleic acids using the polymerase chain reaction (PCR) process. The user interface includes a touchscreen with a graphical display that shows the time, status, and temperature for each step in a PCR run. A touchscreen keypad allows you to enter information into fields on the display screen. Equipment has a simple lid design, which is easy to use and designed to help protect your samples. The equipment is compatible with the Applied Biosystems™ Thermal Cycler Fleet Control Software. Key features and benefits of the thermal cycler are optimizing your PCR with the help of the Veriflex™ blocks (which provide 3 independent temperature zones, for precise temperature control in each step of PCR process), remote access to the system through a mobile application PCR Essentials, transfering data through USB port and simulation modes library, which allows us to run an experiment using the same ramp rates.
Location: ELIXIR-SILVR (KRVS) unit.
 The VeritiPro™ Thermal Cycler (Thermo Fisher Scientific, Inc.) is a thermal cycler, specifically designed for the amplification of nucleic acids using the polymerase chain reaction (PCR) process, with advanced connectivity and temperature control technology. Equipment is compatible with the Applied Biosystems™ Thermal Cycler Fleet Control Software and can be controlled via a touchscreen displaying the time, status and temperature of each PCR process step. The VeritiPro™ Thermal Cycler allows optimizing PCR with the help of the VeriFlex™ blocks (which allows for six zones of independent temperature control through PCR workflow), remote access to the system through a mobile application Connect and simulation modes library, which allows experiment runs using the same ramp rates.
The VeritiPro™ Thermal Cycler (Thermo Fisher Scientific, Inc.) is a thermal cycler, specifically designed for the amplification of nucleic acids using the polymerase chain reaction (PCR) process, with advanced connectivity and temperature control technology. Equipment is compatible with the Applied Biosystems™ Thermal Cycler Fleet Control Software and can be controlled via a touchscreen displaying the time, status and temperature of each PCR process step. The VeritiPro™ Thermal Cycler allows optimizing PCR with the help of the VeriFlex™ blocks (which allows for six zones of independent temperature control through PCR workflow), remote access to the system through a mobile application Connect and simulation modes library, which allows experiment runs using the same ramp rates.
Location: ELIXIR-SILVR (KRVS) unit.
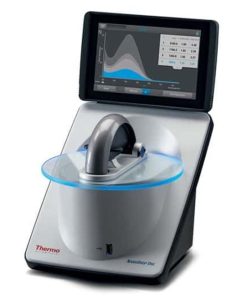 NanoDrop™ One (Thermo Fisher Scientific, Inc.) is a compact, stand-alone UV-VIS spectrophotometer developed for micro-volume analysis of a wide variety of analytes. It allows direct measurements from 1 to 2 µL of sample with auto-range pathlength technology and has embedded applications for DNA, RNA, and protein quantification, providing full spectral data and purity ratios (260/280, 260/230). The equipment is compatible with Thermo Scientific™ Security Suite Software. The software identifies impurities in samples, can differentiate DNA from RNA, and delivers true analyte concentrations. Expanded dynamic range allows measuring up to 27,500 ng/µL dsDNA or 400 mg/mL IgG with no prior dilution. NanoDrop One spectrophotometer features easy data transfer to PC or network via Wi-Fi, USB, or ethernet. Ergonomic touchscreen design and auto-measure capability allows fast and easy operation.
NanoDrop™ One (Thermo Fisher Scientific, Inc.) is a compact, stand-alone UV-VIS spectrophotometer developed for micro-volume analysis of a wide variety of analytes. It allows direct measurements from 1 to 2 µL of sample with auto-range pathlength technology and has embedded applications for DNA, RNA, and protein quantification, providing full spectral data and purity ratios (260/280, 260/230). The equipment is compatible with Thermo Scientific™ Security Suite Software. The software identifies impurities in samples, can differentiate DNA from RNA, and delivers true analyte concentrations. Expanded dynamic range allows measuring up to 27,500 ng/µL dsDNA or 400 mg/mL IgG with no prior dilution. NanoDrop One spectrophotometer features easy data transfer to PC or network via Wi-Fi, USB, or ethernet. Ergonomic touchscreen design and auto-measure capability allows fast and easy operation.
Location: ELIXIR-SILVR (KRVS) unit.
 The Invitrogen Qubit 4 (Thermo Fisher Scientific Inc.) is accurate, specific, sensitive, simple and quick fluorometer for measuring the concentration of DNA, RNA, or protein in a single sample. It can also be used to assess RNA integrity and quality. It requires as little as 1 – 10 µL of sample, even if highly diluted. The user interface includes a touchscreen, which allows you to easily navigate through the various options, with results displayed in just a few seconds. The equipment offers an onboard Reagent Calculator to help in preparing your working solution The Qubit 4 Fluorometer will save the data for up to 1,000 samples, and you can transfer the data to a Thermo Fisher Connect Cloud account via WiFi, or to a computer via a USB drive or cable.
The Invitrogen Qubit 4 (Thermo Fisher Scientific Inc.) is accurate, specific, sensitive, simple and quick fluorometer for measuring the concentration of DNA, RNA, or protein in a single sample. It can also be used to assess RNA integrity and quality. It requires as little as 1 – 10 µL of sample, even if highly diluted. The user interface includes a touchscreen, which allows you to easily navigate through the various options, with results displayed in just a few seconds. The equipment offers an onboard Reagent Calculator to help in preparing your working solution The Qubit 4 Fluorometer will save the data for up to 1,000 samples, and you can transfer the data to a Thermo Fisher Connect Cloud account via WiFi, or to a computer via a USB drive or cable.
Location: ELIXIR-SILVR (KRVS) unit.
 The 2100 Bioanalyzer system (Agilent Technologies, Inc.) is an established automated electrophoresis tool for the sample quality control of biomolecules. The instrument, together with the 2100 Expert Software, provides highly precise analytical evaluation of various samples types in many workflows, including next generation sequencing (NGS), gene expression and biopharmaceutical. Digital data is provided in a timely manner and delivers objective assessment of sizing, quantitation, integrity and purity from DNA, RNA, and protein samples. Minimal sample volumes are required for an accurate result and the data may be exported in many different formats for ease-of-use. The instrument is compliant with European RoHS directives.
The 2100 Bioanalyzer system (Agilent Technologies, Inc.) is an established automated electrophoresis tool for the sample quality control of biomolecules. The instrument, together with the 2100 Expert Software, provides highly precise analytical evaluation of various samples types in many workflows, including next generation sequencing (NGS), gene expression and biopharmaceutical. Digital data is provided in a timely manner and delivers objective assessment of sizing, quantitation, integrity and purity from DNA, RNA, and protein samples. Minimal sample volumes are required for an accurate result and the data may be exported in many different formats for ease-of-use. The instrument is compliant with European RoHS directives.
Location: ELIXIR-SILVR (KRVS) unit.
Laboratory for single cell analysis, location Ljubljana

The LSM 900 confocal microscope allows the study of the three-dimensional distribution of fluorescently labeled molecules in cells and tissues. It is adapted to work with in vitro systems such as cell and tissue cultures, organoids and tissue slices.
The confocal system includes a chamber for maintaining cells at an adjustable temperature and gas concentration (CO2 and O2), which allows long-term experiments on living cells and the study of cell-physiological processes in these cells.
Location: UL MF (KRZS).

The Chromium Controller enables Chromium Single Cell (SC) and Linked-Reads Genomics Solutions. Chromium SC analyses include gene expression, epigenetics (ATAC), cell surface proteins analysis, immune profiling and targeted gene expression on a single-cell level.
The Chromium platform, powered by Next GEM technology, enables integrated analysis of SC at a massive scale. The key to this technology is the ability to generate tens of thousands of single-cell partitions, each containing a unique identifying barcode for downstream analysis.
Location: UL MF (KRZS)
 GridION is a third-generation sequencer from Oxford Nanopore Technologies, which enables long DNA, RNA or cDNA fragments sequencing. Nucleic acid sequence identification is based on the change in electric current during the passage of DNA/RNA through a protein pore.
GridION is a third-generation sequencer from Oxford Nanopore Technologies, which enables long DNA, RNA or cDNA fragments sequencing. Nucleic acid sequence identification is based on the change in electric current during the passage of DNA/RNA through a protein pore.
Sequencing can take place on 5 flow cells simultaneously, on which up to 150Gb can be read in total, thus enabling the sequencing of genomes and transcripts of all types of organisms, including humans. Nanopore technology enables sequencing and assembly of as yet unknown genomes. The computer capacity of GridION enables base-calling in real time and thus enables monitoring and control of the sequencing process.
Location: UL MF (KRZS).
Other laboratories
UM
 Preparative flash/HPLC chromatograph puriFlash® 5.250 is used for the isolation of pure compounds from complex mixtures, e.g. natural extracts. It enables isocratic, as well as gradient (up to 4 solvents) separations that are performed either at low pressures (flash chromatography) or high pressures (up to 250 bar – preparative HPLC). The presence of compounds in the eluent is detected by DAD and ELSD detector. Because the system is equipped with an autosampler and fraction collector, work with it is highly automated.
Preparative flash/HPLC chromatograph puriFlash® 5.250 is used for the isolation of pure compounds from complex mixtures, e.g. natural extracts. It enables isocratic, as well as gradient (up to 4 solvents) separations that are performed either at low pressures (flash chromatography) or high pressures (up to 250 bar – preparative HPLC). The presence of compounds in the eluent is detected by DAD and ELSD detector. Because the system is equipped with an autosampler and fraction collector, work with it is highly automated.
The instrument, among other possible usages, enables isolation of pure compounds from natural extracts and therefore enables studies, which would elucidate connections between the structure and function of natural compounds. The experimental results, obtained on pure compounds, will also be explained by methods of computational chemistry.
Till now, the method for isolation of xanthohumol from hop extract was developed, while the isolation of sufficient quantity for testing the antibacterial activity is in progress.
Location: UM, FKKT (KRVS)
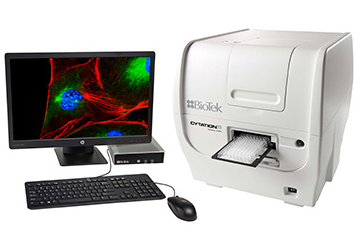 CELL MAPPING SYSTEM AND AUTOMATED MICROSCOPY – CYTATION 5, Manufacturer BioTek (supplier Kemomed, d.o.o.)
CELL MAPPING SYSTEM AND AUTOMATED MICROSCOPY – CYTATION 5, Manufacturer BioTek (supplier Kemomed, d.o.o.)
The cell imaging system enables automated digital microscopy, compatibility with microtiter plates, microscope slides, Petri dishes, bottles for culturing cell culture hemocytometers, allows work with living cells (incubation up to 65 ° C, shaking, optionally also O2 and CO2 control system). Fast cooling after incubation or maintenance of stable ambient temperature (optional additional pellet cooler) should be used, with optional addition use of two reagent injectors, for automatic engineering of reagents during imaging or measurement, and optional possibility of integration with an automated system for simultaneous incubation of up to 8 microtiter plates it is possible to upgrade the system in a multifunctional system that allows the measurement of absorbance, fluorescence, luminescence, fluorescence polarization, and alpha detection.
The system supports the user by enabling: the creation of histograms for data obtained and cell analysis, use of masking for analysis of cytoplasm of cells or whole cells, analysis of intracellular objects, spot counting, analysis of subpopulations with custom-created criteria, use of dynamic thresholds sensitivity in cell mask analysis, production of z-projections, imaging of 3D cell cultures, monitoring of cell migrations, invasions, apoptosis, cell viability, monitoring of fast kinetics, imaging of living cells, and whole organisms (image gluing). The system allows statistical processing of images, automatically identifies patterns for painting (i.e. hit-picking function).
The software also comes with image capture and analysis. The tool is easy to use for users to be supported throughout the retrieval workflow to data processing in an independent user interface. The tool enables the conversion of raw data into images into results, automated image capture, data processing, and analysis in an image retrieval system in ready-to-publish databases.
 HPLC chromatograph Vanquish Core® is used for qualitative and quantitative analysis of various samples. Instrument is equipped with DAD (PDA) detector, which enables determination of concentration of various compounds with chromophore, as well as estimation of peak purity.
HPLC chromatograph Vanquish Core® is used for qualitative and quantitative analysis of various samples. Instrument is equipped with DAD (PDA) detector, which enables determination of concentration of various compounds with chromophore, as well as estimation of peak purity.
Instrument is used in isolation of various natural compound from crude natural extracts. It is used for determination of various compounds concentrations in crude natural extracts which are used as a starting material for isolation of pure compounds. Furthermore, during the isolation process the instrument is used for determination of concentrations and purity of compound of interest in fractions, obtained using preparative chromatography. Finally, it is used in determination of chromatographic purity of isolated compounds.
Successful application: The instrument was recently used in support of isolation of xanthohumol from hop and vescalin and castalin from chestnut wood.
Other research equipment
KI
- Cryo-transmission electron microscope with basic IKT support
- ARC High performance computing center
KIS
- Hyperspectral cameras:
- The two pushbroom imaging spectrometers; HySpex VNIR (spectral range 400–988 nm) and SWIR (950–2500 nm) (Norsk Elektro Optikk AS),
- Multispectral camera 5 bands, RedEdge (Micasense)
- Several qPCR machines (Thermofisher, Roche) and Hamilton pipetting robot
- Droplet digital PCRs QX100 (BioRad)
- Chamber digital PCR BioMark HD (Fluidigm)
UKCLJ
CISLD
- NGS sequencers (Illumina Miseq, ONT MinION)
- NGS library prep infrastructure (enrichment, WGS, RNASEQ, ChIPSeq, long-reads libraries, miRNA-SEQ, Epi-SEQ)
- cfDNA/RNA infrastructure
- Cell Lab
- Brightfield and Flourescent Microscopy Infrastructure
UL BF
- NGS sequencing machine IonProton™ including all necessary equipment
- Sanger sequencing machine ABI Prism 3130XL with 16 capillaries
- Equipment for DNA/RNA/protein analysis (PCR, qPCR, electrophoresis, gel documentation system, samples quantification)
- Linux server with 12 cores, 170 GB RAM, 18 TB disk space
- Software: CLC Genomics Workbench, CLC Genomics Server, Codon Code Aligner, BLAST2GO command line
UL MF
CFGBC
- CFGBC also offers to ELIXIR-SI partners high-throughput and other laboratory equipment
- Genome and Transcriptome analyses
• High density microarrays
• Next Generation Sequencing - Metabolomics (shared, at Vrazov trg 2)
• high performance liquid chromatography with mass spectrometry - Cell culture lab and functional analyses
UL VF
- Ion Torrent PGM sequencer with Ion OneTouch2 (Ion Torrent, Thermo Fischer Scientific)
- Ultrasonicator Covaris M220 Focused
- 1 x system for the evaluation of DNA, RNA and proteins LabChip GX (Caliper – Perkin Elmer)
- Dell PowerEdge R730 Server
- MALDI-TOF mass spectrometer (Bruker Daltonics Inc.)
- 2 x real time PCR instrument ABI PRISM 7500 Real Time PCR System (Applied Biosystems)
- 1 x real time PCR instrument Rotor-Gene Q (Qiagen)
- 2 x real time PCR instrument (LightCycler 1.2, 2.0 LightCycler, Roche)
- 1x realtime PCR instrument QuantStudio 5 Real-Time PCR Systems (Applied Biosystems)
- 1 x digital PCR 3D QS (Applied Biosystems)
- capillary sequencer (ABIPRISM 310, Applied Biosystems)
- Several PCR instrument (Biometra, AB-Thermo Fisher)
- 1 x system for gel documentation GenGenious (Syngen)
- 1 x capillary electrophoresis QIAXCEL (QIAGEN)
- Ultracentrifuge cooling (Beckman)
- 1x Kingfisher automated nucleic acid extraction instrument
UM
- Computer cluster KROP1
- Computer cluster KROP2
- 2x UV/Vis spectrophotometer
| Oprema | Nabavna vrednost (EUR) | Navedi tip opreme (distribuirana; mobila; na lokaciji; virtualna) | Raziskovalno področje | Vir sofinanciranja iz javnih sredstev (evropska kohezijska sredstva; drugi javni viri) | Opis postopka dostopa do opreme - (čas; največ 5 stavkov) | Namembnost opreme in dodatne informacije (največ 5 stavkov); | Cena za uporabo raziskovalne opreme za izučenega uporabnika (v EUR/uro) | Letna stopnja izkoriščenosti v % v pretek. koled. letu | Spletna stran RO (predstavitev opreme; pogoj dostopa; cenik) | Številka | Mesečno povprečno število uporabnikov | Mesečna povprečna stopnja izkoriščenosti v % |
|---|




